Guinier analysis¶
Recall Guinier’s approximation at low-q: \(I(q)\approx I(0) \exp(-R_g^2 q^2 /3)\).
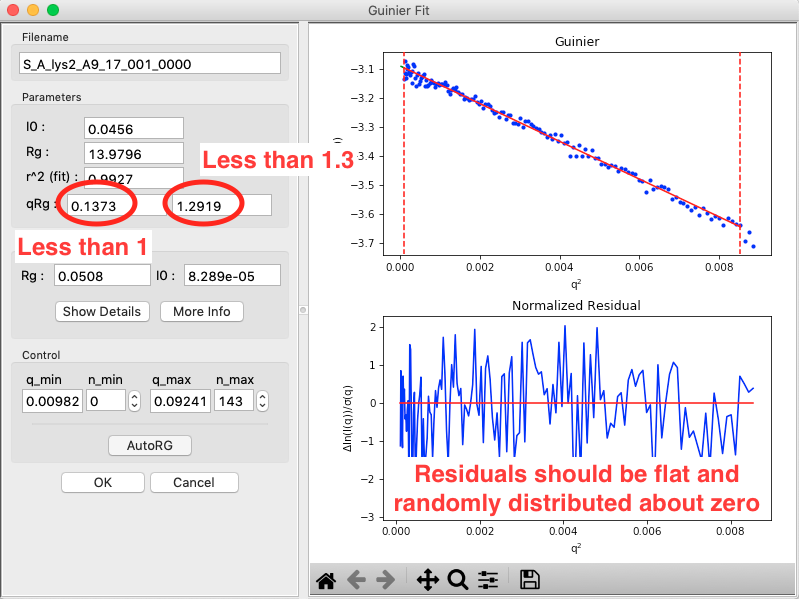
Rg and I(0) can be determined by performing a linear fit in the Guinier plot (a plot of \(\ln(I)\) vs. \(q^2\)). The fitting region should normally have \(q_{max}R_g<1.3\) for globular proteins. This fitting region is called the “Guinier region.”
In RAW, right click (ctrl click on macs without a right mouse button) on the subtracted GI scattering profile in the Manipulation list and select “Guinier fit”. In the plots on the right, the top plot shows you the Guinier plot and the fit, while the bottom plot shows you the residual of the fit.
- Note: RAW automatically tries to find the best Guinier region for you when the Guinier window is opened for the first time.
- Note: The Rg value is in Angstroms, while the two \(qR_g\) boxes give, left to right, \(q_{min}R_g\) and \(q_{max}R_g\) respectively.
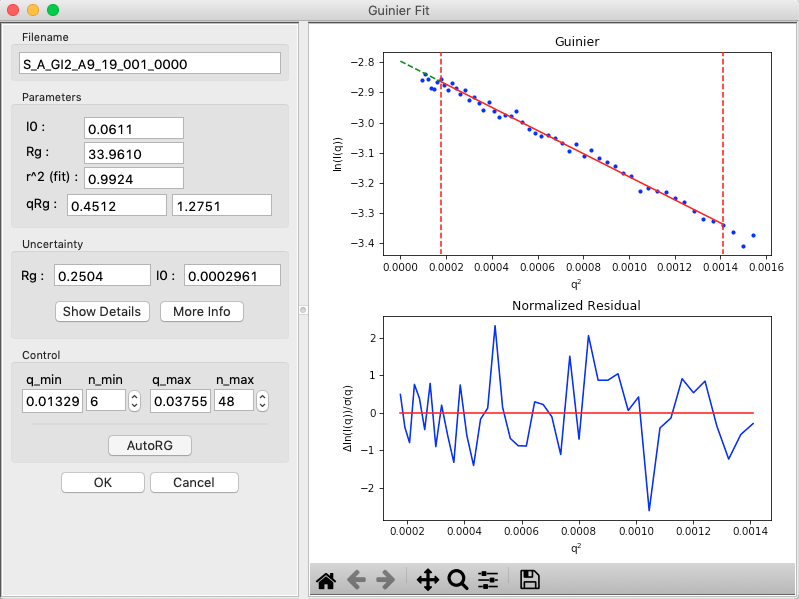
In the “Control” panel, you’ll see that n_min is now 6. This means RAW has cut off the first six points of the scattering profile in the fit. Use the arrow buttons next to the n_min box to adjust that to zero. Check whether the Rg changes.
In the “Parameters” panel, note that \(q_{max}R_g\) is only ~1.26. Recall that for globular proteins like GI, it is typical to have \(q_{max}R_g\) ~1.3. Adjust n_max until that is the case, watching what happens to the Rg and the residual.
- Question: The literature radius of gyration for GI is 32.7 Å. How does yours compare?
RAW also provides an estimate of the uncertainty in both the Rg and I(0) values for the Guinier fit, shown in the Uncertainty section.
- Note: This is the largest of the uncertainties from the fit (standard deviation of fit values calculated from the covariance matrix), and either the standard deviation of Rg and I(0) across all acceptable intervals found by the autorg function or an estimated uncertainty in Rg and I(0) based on variation of the selected interval start and end points.
Click the “OK” button to keep the results.
- Checkpoint: If you now select the GI scattering profile, in the information panel at the top you should see the Rg and I(0) that you just found.
- Note: Clicking the “Cancel” button will discard the results.
Repeat the Guinier analysis for lysozyme.
- Try: Increase qmin and/or decrease qmax to verify that the Rg does not change significantly in the Guinier region.
- Tip: If you hover your mouse cursor over the info icon (just left of the target icon) for a given scattering profile it should show you the Rg and I(0) of your Guinier analysis.