Molecular weight analysis¶
RAW provides four forms of molecular weight analysis:
- Referencing I(0) to that of a known standard
- From the volume of correlation using the method of Rambo and Tainer
- From the adjusted Porod volume using the method of Fisher et al.
- From the value of I(0) on an absolute scale.
In RAW, right click on the subtracted GI scattering profile in the manipulation panel and select “Molecular weight.” At the top of the panel is a description of the methods used, and the results of your Guinier fit. All four methods require a good Guinier fit, so you can use that button to redo the fit if necessary. In the lower part of the panel, the results of the four estimates for MW are shown.
- Note: Neither the I(0) Ref. MW panel nor the Abs. MW panel should be reporting a MW.
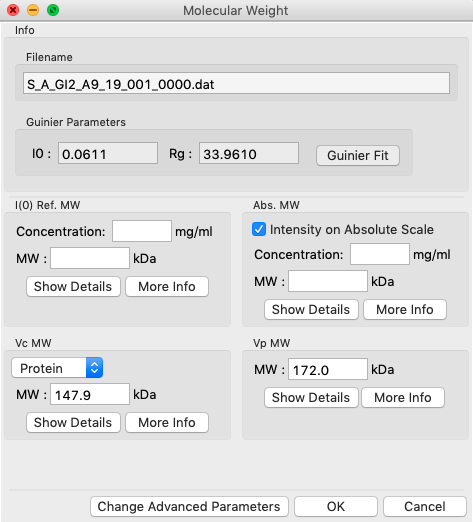
In either concentration box, enter the sample concentration of 0.47 mg/ml. Notice that you now get results from all four methods of MW calculation.
- Question: The expected MW value for GI is 172 kDa. How do your results compare?
Click on the “Show Details” button for the Vc MW panel. You should see a graph, which shows the integrated area of \(qI(q)\) vs. q. For this method to be accurate, this value needs to converge at high q.
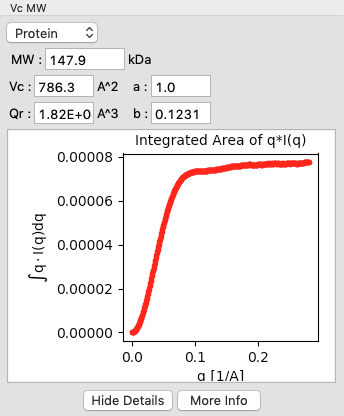
Click the “OK” button to save your analysis.
- Note: The “Cancel” button discards the analysis.
Repeat the MW analysis for the lysozyme sample, which had a concentration of 4.27 mg/ml. The expected MW of lysozyme is 14.3 kDa.
- Question: Does the Vc method work for the lysozyme data?